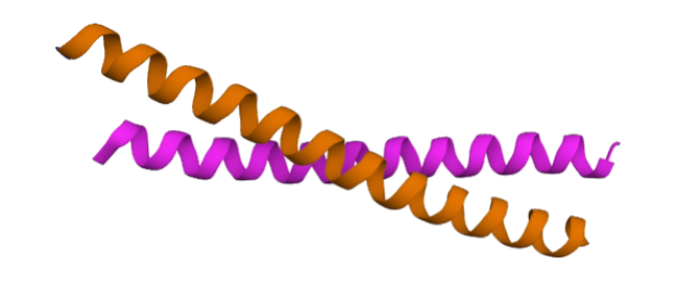
 The CoCoNat web server
The CoCoNat web server
Submission of a single sequence
From the CoCoNat home page, a single sequence in fasta format can be submitted for coiled-coil analysis
by copying the text in the panel or uploading a FASTA file, and pressing the button "Submit". Valid inputs include an ID line starting with a '>'
character, followed by the protein sequence, that can have between 40 and 700 valid residues (including the 20 standard amino acids ARNDCQEGHILKMFPSTWYV,
non-standard residues UZB, and undetermined residue X).
An example sequence can be loaded clicking on the Example button.
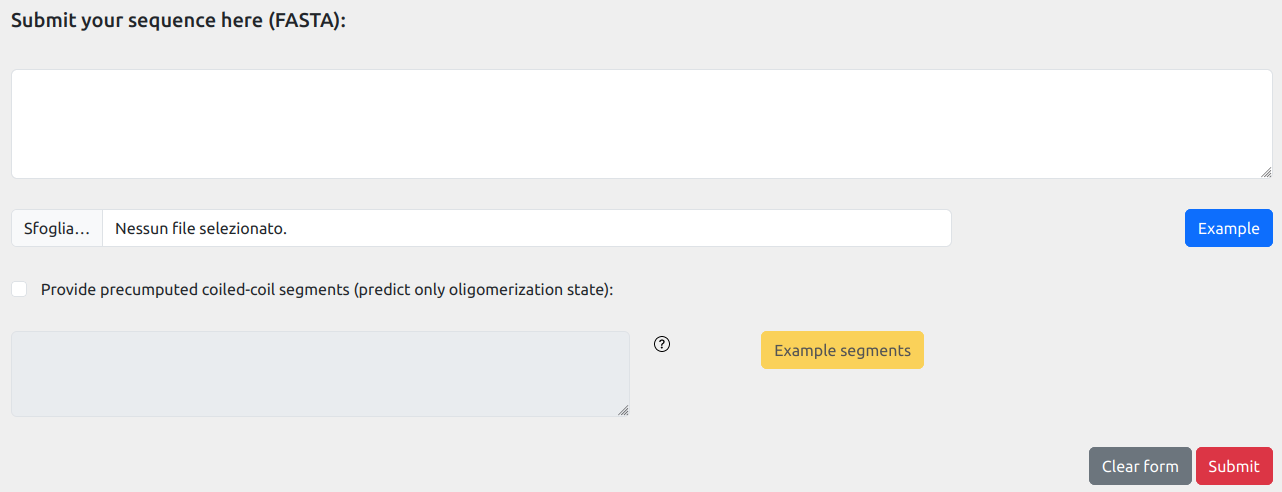
By default, CoCoNat works in abinitio mode: given an input sequence, it firstly detects the coiled-coil segments
and then performs oligomerization state prediction on predicted segments.
If you already have coiled-coil segments (either experimentally verified or predicted), you can also run CoCoNat
in state prediction mode only, providing the annoatation of available segments along the input sequence.
To do so, you need to check the "Provide precomputed coiled-coils segments" flag. By this, a new text area will appear
where you can provide your annotation.
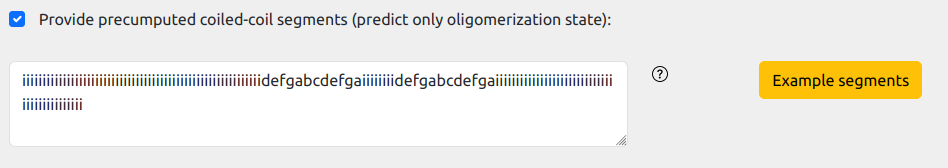
The format for the is straightforward, namely a string of the same length of the protein,
specifying, for each residue position, one of eigth possibile labels: abcdef for coiled-coils heptad repeat registers, i for non-coiled coils residues.
For example, for the sequence:
>P16087
MGNGQGRDWKMAIKRCSNVAVGVGGKSKKFGEGNFRWAIRMANVSTGREPGDIPETLDQLRLVICDLQERREKFGSSKEIDMAIVTLKVFAVAGLLNMTVSTAAAAENMYSQMGLDTRPSMKEAGGKEEGPPQAY
a possible coiled-coil annotation string, iuncluding two distinct coiled-coil segments, can be:
iiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiidefgabcdefgaiiiiiiiidefgabcdefgaiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiii
IMPORTANT: State prediction mode is only available if you have precise heptad-repeat register annotation, since this piece of information is used by CoCoNat to construct the input for predicting the oligomerization state.
An example sequence can be loaded clicking on the Example segments button.
Result page
A typical CoCoNat result looks like the following:
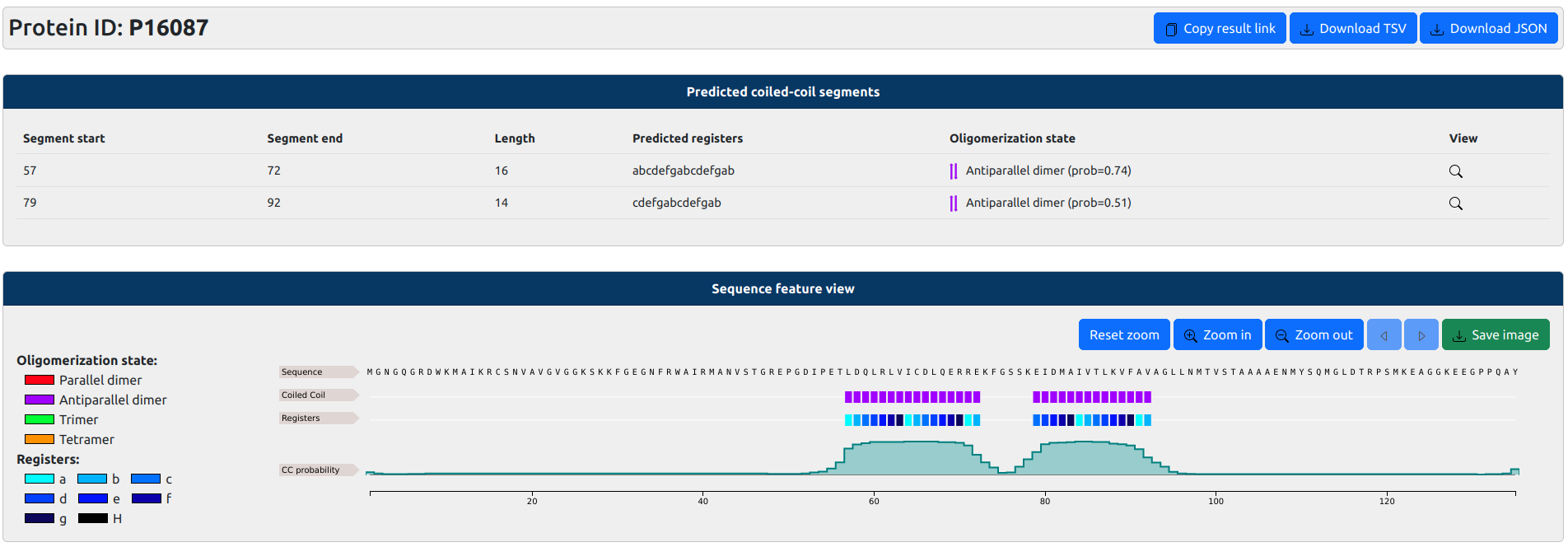
On top, protein ID is shown, as well as buttons to copy the results page (for future access), or download results in
TSV or JSON formats.
Then a panel reports information about predicted segments in tabular format, including: segment start and end, length,
heptad repeat register annotation, pedicted oligomerization state. Clicking on the

A second panel visualize the predicted coiled-coil segments, registers and oligomerization state along the sequence using
an inteactive feature viewer. Three tracks are shown corresponding to coiled-coil segment annotation (coloured by oligomenrization state), corresponding
heptad repeat register annotation, and coiled-coil probability.
If you hover the mouse over the graphs, the position of the residue will be shown.
If you select a rectangular area, you can zoom on a part of the sequence. You can always right click on
the graph to reset the zoom back to normal. Alternatively, you can make use of the
buttons on the top-right of the viewer to zoom-in, zoom-out, move along the sequence or
take a screenshot of the selected area.